🧬 Exploring nf-core RNA-seq
On day four of the Applied Bioinformatics course, we worked with the “nf-core” framework to search for and run a standardized RNA-seq pipeline.

We chose the nf-core/rnaseq pipeline, this is a well-documented and modular workflow built on Nextflow that handles everything from quality control to read alignment and quantification.
⚙️ How the Pipeline Works
Here’s an overview of the pipeline architecture:
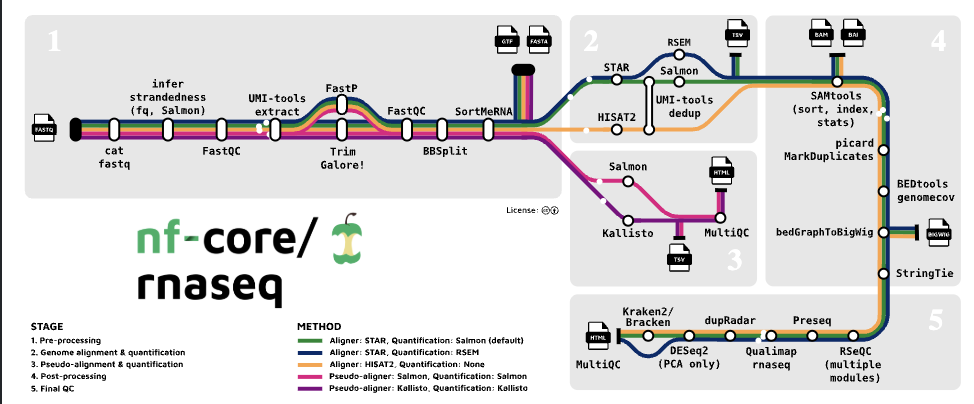
Each box represents a process in the workflow (e.g., trimming, alignment, quantification), all managed via Nextflow. It also includes MultiQC for aggregating all the QC reports at the end.
This was a nice practice on how to use nf-core.