Change of Plans !!!!
On day 4 of the Applied Bioinformatics course, we were scheduled to work hands-on with Nextflow pipelines. However, due to technical issues with the HPCN cluster, we couldn’t access the environment.
Instead, the day turned into a discussion-session focused on:
- Understanding the structure and logic behind Nextflow pipelines
- Reflecting on how we use AI in bioinformatics workflows
⚙️ What is Nextflow?

Nextflow is a workflow management system designed to turn computational scripts into modular, reproducible processes that can run independently and in parallel.
The main building blocks of a Nextflow pipeline are:
Processes:
These are the individual tasks, which can be in any language (bash, Python, R, etc.) and runs independently.Channels:
These act as the data pipes between processes, passing files, variables, or inputs.
We spent time understanding: - Which files are needed to run a pipeline - How to manage environments using Pixi
🔁 Channels and Processes
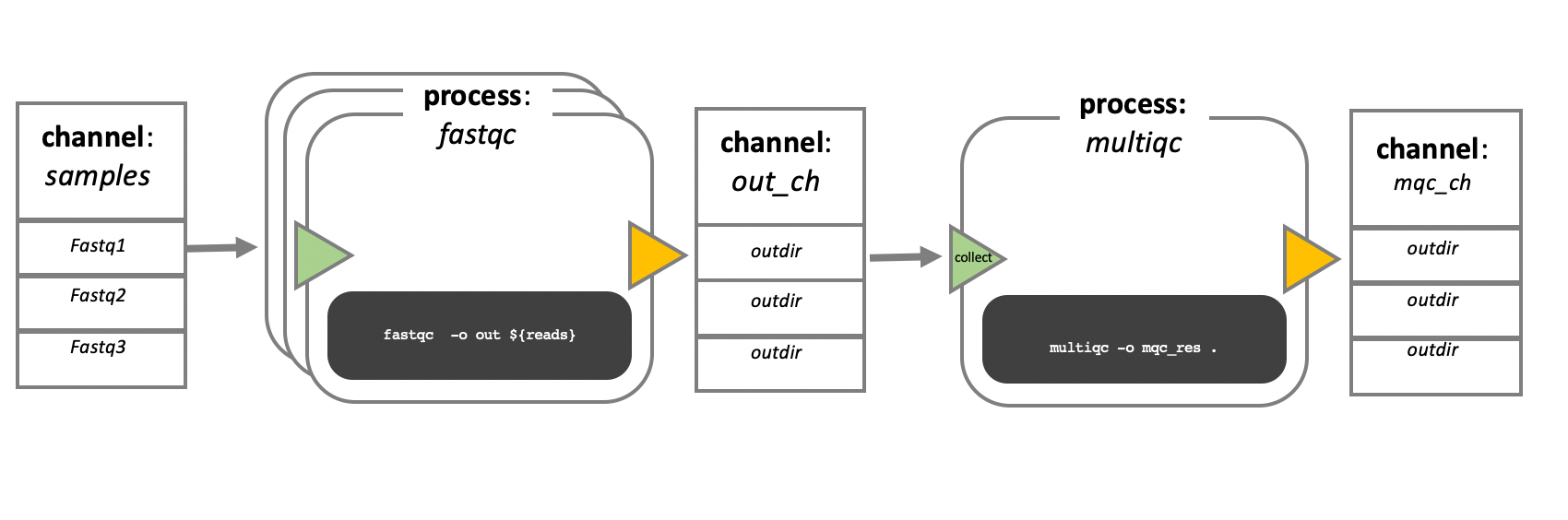
One key idea is that processes don’t directly depend on each other — they communicate through channels.
You define the what (your process), and Nextflow handles the when and how based on available inputs.
This makes pipelines scalable, portable, and easy to parallelize —> especially important when working with large data on HPC clusters or cloud systems.
🧠 Reflections
Although we couldn’t run pipelines today, the discussion helped clarify some important questions I had about workflow design and reproducibility.
I also liked the conversation around how AI tools are increasingly becoming part of our daily practice as bioinformaticians. We also gave examples of how we used them.
✅ Key Takeaways
- Nextflow = scripts + processes + channels
- Each process can be written in any language — it’s fully modular
- Channels are the communication system — they connect processes with data
- Workflow tools like Pixi simplify environment management for reproducibility
- Even when we can’t run code, talking through design and structure is essential for deep understanding
📌 This day we also had the course dinner. This was a very nice oportunity to get to know everyone a bit better and enjoy a nice dinner. Thanks to Samuel and Amrei for making this happen!